“We want to make biological data science accessible to the world.” That is the mission statement of BioLib, a Copenhagen-based startup founded in 2019 to accelerate the life sciences using software. The team is capturing algorithms being developed through GitHub to test ideas in biological sciences, benchmarking them, and then making them widely available for others in the field. However, the team at BioLib isn’t just doing the technical groundwork of building a secure and scalable system for sharing and benchmarking these algorithms – they are equally busy trying to grow the community of algorithm developers and benchmarks to drive innovation in the space of biological data science.
Hackathons for a global bioinformatics community
In early 2020 BioLib co-founders Jeppe Hallgren and Jørn Emborg joined forces with a local computational biology society, CBioVikings, planning a Copenhagen based in-person hackathon in April. Then the pandemic hit, so the team had to adapt, deciding to go online. This turned out to be a blessing in disguise, allowing to open up the event for a much bigger global community.
Building on that success, in April 2021, BioLib and its partners hosted the second Copenhagen Bioinformatics Hackathon. The team gathered nine challenges from various Danish biopharma companies and from research groups at a range of universities, including Oxford, the Technical University of Denmark and more. With 500+ applicants from all over the world, the event was heavily over-subscribed. BioLib’s Jørn Emborg noted, “It’s really awesome that so many people are ready to spend their weekend working on the challenges! Most people, of course, also do it for the fun of it, but I think a lot of the motivation is also to learn new skills. Some people who know about biology may be great programmers, but they haven’t worked much with machine learning in practice. The hackathon was a chance for them to explore that.”
Balancing collaboration and competition
Inspired by the CASP, the biannual protein folding competition, the BioLib team decided to implement a benchmarking system to spice things up with a bit of “good-natured competition.” The benchmarking system was set up such that participants could easily push models directly from GitHub to BioLib’s zero-knowledge benchmarking servers. Unlike at former hackathons, the benchmarking system enabled teams to see how their work was faring against that of their peers’ on the real-time leaderboard. Having the leaderboard proved a big motivator for the participants. “Earlier, people had lost steam around Saturday evening because they had been going for 24 hours and had no feedback. But this time around, it was a completely different mindset. It was like, ‘We’re top three – almost there!’ And they would just keep going,” commented Jeppe. While Jørn and the team were happy to use a bit of friendly competition to motivate the participants, they emphasized that it was important to them that they, “didn’t want this to get too competitive. The whole point in the first place is for everyone to meet new people and learn from each other. Collaboration is clearly the way forward!”
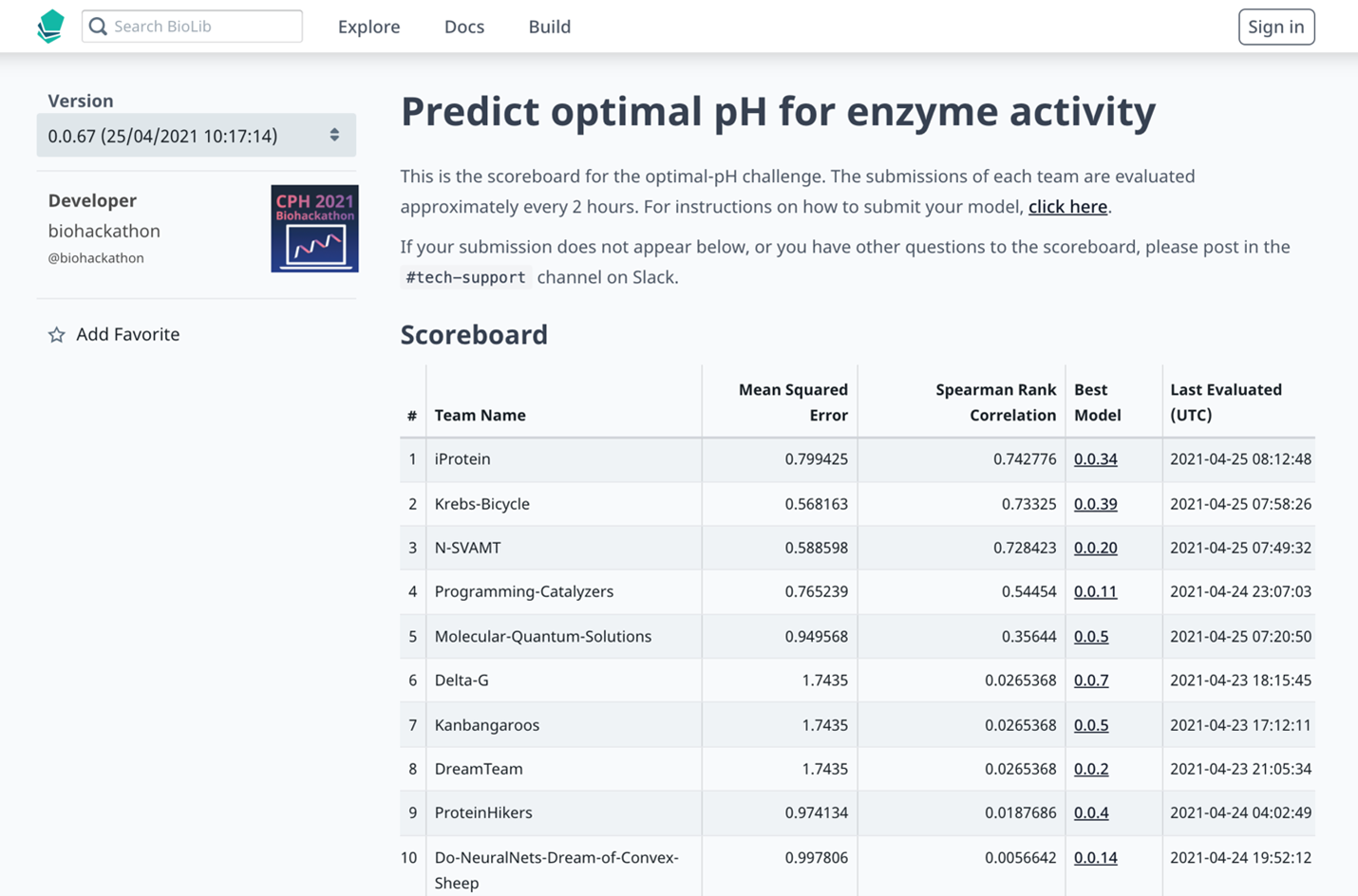
The live scoreboard for one of the challenges at the Copehagen Bioinformatics Hackathon 2021.
Using GitHub for Automatic Benchmarking
It was important to get the participants a good start. Jeppe noted, “We created some template code that parsed the data, and a baseline model that made some simple predictions. At the start of the hackathon, all you had to do was to fork the repository to your team’s GitHub account, and then right away, your team had a baseline model that actually worked on real data.” The team also found GitHub Actions to be very useful in simplifying the workflows around the benchmarking. GitHub Actions enables you to identify an action, event or workflow that runs every time you push a new update or write a new line of code. For the hackathon, they defined the action as pushing the results of the code to the BioLib benchmarking server and updating the leaderboard. “Given the short amount of time, we were concerned that this benchmarking system would be too complex to get working. But GitHub Actions made setting up the leaderboard very straightforward,” Jeppe commented. The setup with pre-configured repositories on GitHub, and automated actions for the benchmarking system, were critical components of scaling the hackathon globally. This allowed the hundreds of participants, each with very different backgrounds and skill levels, to make meaningful progress in just two days. Jeppe added, “We tried to make everything pre-configured, so the hackathon teams could just focus on creating the best possible solution for the problem at hand.”
Going forward
As the BioLib team thinks about the future, they intend to both scale and refine their approach. “It was great with so many applicants, and we want make sure these challenges are available to anyone that wants to participate. To handle more submissions, we think that it will work better if we have people coming together for a single challenge rather than having an event with nine different challenges running at the same time” suggested Jørn. He added that “a lot of participants told us they think that a longer event would be better, so we will experiment with challenges running over several weeks, where the teams will have more time to improve their solutions.”
The BioLib team’s ambition is to continue growing the community, bringing together more hackers, biopharma companies, and universities. They hope the hackathons, in the long run, will encourage development of new and better algorithms and that the benchmarking system they are building will help researchers gauge which algorithms are actually the best ones for a given problem. “For example, perhaps a researcher wants to know, ‘What is the optimal pH for this enzyme?’ The dream is that they can just go to the benchmark page and immediately find out what the best algorithm for that task is, and then run it on their enzyme. It’s just a great way to create scientific progress and awareness, and that’s really what the world needs,” Jeppe said.












